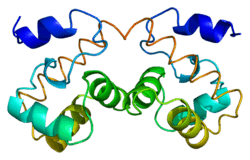Barrier to autointegration factor 1
Barrier-to-autointegration factor is a protein that in humans is encoded by the BANF1 gene.[5][6] It is a member of the barrier-to-autointegration factor family of proteins.
Function
The protein encoded by this gene was identified by its ability to protect retroviruses from intramolecular integration and therefore promote intermolecular integration into the host cell genome. The endogenous function of the protein is unknown. The protein forms a homodimer which localizes to the nucleus and is specifically associated with chromosomes during mitosis. This protein binds to DNA in a non-specific manner and studies in rodents suggest that it also binds to lamina-associated polypeptide 2, a component of the nuclear lamina.[6] It also associates with the LEM Domain containing proteins LAP2, Emerin, and MAN1. The protein's DNA binding ability is modulated by ATP concentration.[7]
Interactions
Barrier to autointegration factor 1 has been shown to interact with Thymopoietin.[8]
Clinical relevance
Mutations in this gene have been shown to cause hereditary progeroid syndrome.[9]
See also
- Retroviral integration
References
- GRCh38: Ensembl release 89: ENSG00000175334 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000024844 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Lee MS, Craigie R (Mar 1998). "A previously unidentified host protein protects retroviral DNA from autointegration". Proc Natl Acad Sci U S A. 95 (4): 1528–33. doi:10.1073/pnas.95.4.1528. PMC 19075. PMID 9465049.
- "Entrez Gene: BANF1 barrier to autointegration factor 1".
- Sridharan S, Kurzawa N, Werner T, Günthner I, Helm D, Huber W, Bantscheff M, Savitski MM (March 2019). "Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP". Nature Communications. 10 (1): 1155. doi:10.1038/s41467-019-09107-y. PMC 6411743. PMID 30858367.
- Furukawa K (August 1999). "LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction". J. Cell Sci. 112 (Pt 15): 2485–92. PMID 10393804.
- Puente XS, Quesada V, Osorio FG, Cabanillas R, Cadiñanos J, Fraile JM, Ordóñez GR, Puente DA, Gutiérrez-Fernández A, Fanjul-Fernández M, Lévy N, Freije JM, López-Otín C (May 2011). "Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome". Am. J. Hum. Genet. 88 (5): 650–6. doi:10.1016/j.ajhg.2011.04.010. PMC 3146734. PMID 21549337.
Further reading
- Goldberg M, Harel A, Gruenbaum Y (2000). "The nuclear lamina: molecular organization and interaction with chromatin". Crit. Rev. Eukaryot. Gene Expr. 9 (3–4): 285–93. doi:10.1615/critreveukargeneexpr.v9.i3-4.130. PMID 10651245.
- Segura-Totten M, Wilson KL (2004). "BAF: roles in chromatin, nuclear structure and retrovirus integration". Trends Cell Biol. 14 (5): 261–6. doi:10.1016/j.tcb.2004.03.004. PMID 15130582.
- Van Maele B, Debyser Z (2005). "HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins". AIDS Reviews. 7 (1): 26–43. PMID 15875659.
- Van Maele B, Busschots K, Vandekerckhove L, et al. (2006). "Cellular co-factors of HIV-1 integration". Trends Biochem. Sci. 31 (2): 98–105. doi:10.1016/j.tibs.2005.12.002. PMID 16403635.
- Lee MS, Craigie R (1994). "Protection of retroviral DNA from autointegration: involvement of a cellular factor". Proc. Natl. Acad. Sci. U.S.A. 91 (21): 9823–7. doi:10.1073/pnas.91.21.9823. PMC 44909. PMID 7937898.
- Dear PH, Bankier AT, Piper MB (1998). "A high-resolution metric HAPPY map of human chromosome 14". Genomics. 48 (2): 232–41. doi:10.1006/geno.1997.5140. PMID 9521877.
- Lynch RA, Piper M, Bankier A, et al. (1999). "Genomic and functional map of the chromosome 14 t(12;14) breakpoint cluster region in uterine leiomyoma". Genomics. 52 (1): 17–26. doi:10.1006/geno.1998.5406. PMID 9740667.
- Cai M, Huang Y, Zheng R, et al. (1998). "Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration". Nat. Struct. Biol. 5 (10): 903–9. doi:10.1038/2345. PMID 9783751.
- Furukawa K (1999). "LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction". J. Cell Sci. 112 (15): 2485–92. PMID 10393804.
- Zheng R, Ghirlando R, Lee MS, et al. (2000). "Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex". Proc. Natl. Acad. Sci. U.S.A. 97 (16): 8997–9002. doi:10.1073/pnas.150240197. PMC 16810. PMID 10908652.
- Umland TC, Wei SQ, Craigie R, Davies DR (2000). "Structural basis of DNA bridging by barrier-to-autointegration factor". Biochemistry. 39 (31): 9130–8. doi:10.1021/bi000572w. PMID 10924106.
- Harris D, Engelman A (2001). "Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro". J. Biol. Chem. 275 (50): 39671–7. doi:10.1074/jbc.M002626200. PMID 11005805.
- Lee KK, Haraguchi T, Lee RS, et al. (2002). "Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF". J. Cell Sci. 114 (Pt 24): 4567–73. PMID 11792821.
- Haraguchi T, Koujin T, Segura-Totten M, et al. (2002). "BAF is required for emerin assembly into the reforming nuclear envelope". J. Cell Sci. 114 (Pt 24): 4575–85. PMID 11792822.
- Segura-Totten M, Kowalski AK, Craigie R, Wilson KL (2002). "Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly". J. Cell Biol. 158 (3): 475–85. doi:10.1083/jcb.200202019. PMC 2173821. PMID 12163470.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Holaska JM, Lee KK, Kowalski AK, Wilson KL (2003). "Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro". J. Biol. Chem. 278 (9): 6969–75. doi:10.1074/jbc.M208811200. PMID 12493765.
- Lin CW, Engelman A (2003). "The Barrier-to-Autointegration Factor Is a Component of Functional Human Immunodeficiency Virus Type 1 Preintegration Complexes". J. Virol. 77 (8): 5030–6. doi:10.1128/JVI.77.8.5030-5036.2003. PMC 152146. PMID 12663813.