Aspergillomarasmine A
Aspergillomarasmine A is an polyamino acid naturally produced by the mold Aspergillus versicolor. The substance has been reported to inhibit two antibiotic resistance carbapenemase proteins in bacteria, New Delhi metallo-beta-lactamase 1 (NDM-1) and Verona integron-encoded metallo-beta-lactamase (VIM-2), and make those antibiotic-resistant bacteria susceptible to antibiotics.[1] Aspergillomarasmine A is toxic to leaves of barley and other plants, being termed as "Toxin C" when produced by Pyrenophora teres.[2]
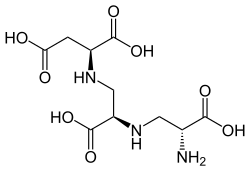 | |
| Names | |
|---|---|
| IUPAC name
(R-(R*,R*))-N-(2-((2-Amino-2-carboxyethyl)amino)-2-carboxyethyl)-L-aspartic acid | |
| Identifiers | |
3D model (JSmol) |
|
| ChemSpider | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C10H17N3O8 | |
| Molar mass | 307.257 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
The molecule is a tetracarboxylic acid with four -COOH groups. One section of the molecule is the amino acid aspartic acid. This has two alanine molecules attached. Aspergillomarasmine B differs in that the last alanine is replaced by glycine.
The crystalline substance was first isolated in 1956, but its name was given until 1965.[3]
In addition to Aspergillus versicolor, aspergillomarasmine A is also produced by the ascomycete Pyrenophora teres where it acts as a toxin in the barley net-spot blotch disease. In P. teres, a biosynthetic precursor of aspergillomarasmine A, L,L-N-(2-amino-2-carboxyethyl)-aspartic acid has also been isolated and found to contribute to the phytotoxic properties of this microbe.[4] This precursor, aspergillomarasmine A itself, and a lactam form (anhydroaspergillomarasmine A) are together termed the marasmines.[2]
Other producers of aspergillomarasmine A include Aspergillus flavus,[3] Aspergillus oryzae,[5] Colletotrichum gloeosporioides, and Fusarium oxysporum.[2]
In mice the LD50 toxic dose of aspergillomarasmine A is 159.8 mg/kg.[6]
Properties
Aspergillomarasmine A takes the form of colourless crystals. The chemical is insoluble in common organic solvents, but can dissolve in water under either basic or strongly acidic conditions.[3]
Anhydroaspergillomarasmine A, a lactam of aspergillomarasmine A, chemically called [1-(2-amino-2carboxyethyl)-6-carboxy-3-carboxymethyl-3-piperazinone], can also be found in Pyrenophora teres. The relative amount of these two toxins is dependent upon the pH of the growth medium, with lower pH favouring the lactam form.[2] The lactam can be hydrolyzed to aspergillomarasmine A by treating it with trifluoroacetic acid.[2]
Aspergillomarasmine A functions as a chelating agent, sequestering Fe3+ ions.[7] It can inhibit endothelin converting enzymes even in the live rat, probably by chelating metals required by metalloproteases.[8]
When heated, aspergillomarasmine A decomposes between 225° and 236 °C. Hydrolysis produces L-aspartic acid and racemic 2,3-diamino-propionic acid. [α]20°D at pH 7 is -48°.[3]
With nitrous acid aspergillomarasmine A is deaminated, and isoserine with aspartic acid is formed.[3]
Titration reveals changes in ionisation at pK 3.5 and 4.5 due to carboxylic acid groups, and pK 9.5 and 10 due to amino groups.[3]
Treatment with ninhydrin shows a purple colour.[3]
References
- King, Andrew M.; Sarah A. Reid-Yu; Wenliang Wang; Dustin T. King; Gianfranco De Pascale; Natalie C. Strynadka; Timothy R. Walsh; Brian K. Coombes; Gerard D. Wright (2014). "Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance". Nature. 510 (7506): 503–506. Bibcode:2014Natur.510..503K. doi:10.1038/nature13445. ISSN 0028-0836. PMC 4981499. PMID 24965651.
- Weiergang, I.; H.J. Lyngs Jørgensen; I.M. Møller; P. Friis; V. Smedegaard-Petersen (2002). "Optimization of in vitro growth conditions of Pyrenophora teres for production of the phytotoxin aspergillomarasmine A". Physiological and Molecular Plant Pathology. 60 (3): 131–140. doi:10.1006/pmpp.2002.0383. ISSN 0885-5765.
- Haenni, A. L.; M. Robert; W. Vetter; L. Roux; M. Barbier; E. Lederer (1965). "Structure chimique des aspergillomarasmines A et B (Chemical structure of aspergellomarasmines A and B)". Helvetica Chimica Acta (in French). 48 (4): 729–750. doi:10.1002/hlca.19650480409. ISSN 0018-019X. PMID 14321962.
- Friis, P; Olsen C.E.; Møller B.L. (15 July 1991). "Toxin production in Pyrenophora teres, the ascomycete causing the net-spot blotch disease of barley (Hordeum vulgare L.)". The Journal of Biological Chemistry. 266 (20): 13329–13335. PMID 2071605.
- Wagman, G.H.; Cooper, R. (1988-12-01). Natural Products Isolation: Separation Methods for Antimicrobials, Antivirals and Enzyme Inhibitors. Elsevier. p. 499. ISBN 9780080858487. Retrieved 27 June 2014.
- Matsuura, Akihiro; Hiroshi Okumura; Rieko Asakura; Naoki Ashizawa; Mayumi Takahashi; Fujio Kobayashi; Nami Ashikawa; Koshi Arai (1993). "Pharmacological profiles of aspergillomarasmines as endothelin converting enzyme inhibitors". The Japanese Journal of Pharmacology. 63 (2): 187–193. doi:10.1254/jjp.63.187. PMID 8283829.
- Barbier, M. (1987). "Remarks on the biological activity of aspergillomarasmine A Fe3+ chelate and other iron transporting phytotoxins with reference to their role in the photodegradation of aromatic amino-acids in infected plant leaves". Journal of Phytopathology. 120 (4): 365–368. doi:10.1111/j.1439-0434.1987.tb00500.x. ISSN 0931-1785.
- Huggins, John P.; Pelton, John T. (1996-12-23). Endothelins in Biology and Medicine. CRC Press. p. 121. ISBN 9780849369759. Retrieved 27 June 2014.