Antibiotic sensitivity testing
Antibiotic sensitivity testing or antibiotic susceptibility testing is the measurement of the susceptibility of bacteria to antibiotics. It is used because bacteria may have resistance to some antibiotics. Knowledge about what antibiotics a bacterium is sensitive to can change the choice of antibiotics from empiric therapy to directed therapy.[1]

Sensitivity testing usually occurs in a laboratory setting, and may be based on culture methods that exposure bacteria to antibiotics, or genetic methods that test to see if a bacterium possesses genes that confer resistance. Culture methods often involve measuring the diameter of the zones of inhibition on agar culture dishes of bacterial growth around paper discs that are impregnated with antibiotics. The minimum inhibitory concentration of the antibiotic can be determined from this.
Uses
In clinical medicine, antibiotics are most frequently prescribed on the basis of a person's symptoms and general guidelines, called empiric therapy.[1] These are based around knowledge about what bacteria cause an infection, and also what antibiotics bacteria may be sensitive or resistant to in a geographical area.[1] For example, a simple urinary tract infections might be treated with trimethoprim/sulfamethoxazole.[2] This is because Escherichia coli is the most likely causative bacterium, and may be sensitive to that combination antibiotic.[2] However, bacteria can be resistant to several classes of antibiotics.[2] This resistance might be because of the type of bacteria,[2] because of resistance following past exposure to with antibiotics,[2] or because resistance may be transmitted from other sources such as plasmids.[3] Antibiotic sensitivity testing provides information about what antibiotics are more likely to be successful and are used in this context to provide information about what antibiotics should be used.[1]
Antibiotic sensitivity testing is also conducted at a population level in some countries as a form of screening.[4] This is to assess the background rates of resistance to antibiotics (for example with Methicillin-resistant Staphylococcus aureus), and may influence guidelines and public health measures.[4]
Methods
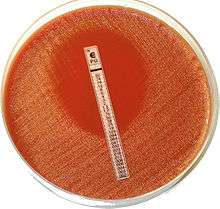
Testing for antibiotic sensitivity usually occurs in a laboratory setting.[5] Once a bacterium has been identified after microbiological culture testing, certain antibiotics are selected for susceptibility testing.[6] Susceptibility testing methods are based on exposing bacteria to antibiotics and observing the response ("phenotypic testing"), or specific genetic tests ("genetic testing").[7] Methods used may be qualitative, meaning a result indicates resistance is or is not present; or quantitative, using a minimum inhibitory concentration to describe the concentration of antibiotic to which a bacterium is sensitive.
Phenotypic methods
Testing based on exposing bacteria to antibiotics uses agar plates or dilution in agar or broth.[8] The selection of antibiotics will depend on the organism grown, and the antibiotics that are available locally.[6]
The disc diffusion method involves selecting a strain of bacteria, and placing it in a growth media.[9] This is also called the Kirby-Bauer method,[10] although modified methods are also used.[11] Small paper discs containing antibiotics are placed onto a plate upon which bacteria are growing. If the bacteria are sensitive to the antibiotic, a clear ring, or zone of inhibition, is seen around the wafer indicating poor growth.[11] Mueller-Hinton agar is frequently used in this antibiotic susceptibility test.[11] Standards exist as to how the testing is performed, and how the test results are interpreted.[6] It is considered the cheapest and most simple of the methods used to test for susceptibility, and is easily adapted to testing newly available antibiotics or formulations.[6] Limitations exist for some types of bacteria, including slow-growing and fastidious bacteria, which cannot be accurately tested.[6]
Gradient methods.[6] The Etest, which uses a plastic strip placed on agar is an example of this.[6] A plastic strip impregnated with different concentrations of antibiotics is placed on a growth medium, and the growth medium is viewed after a period of incubation.[6] Multiple strips for different antibiotics may be used.[6] This type of test is considered a diffusion test.[12]
Agar and Broth dilution methods.[6] In these methods, bacteria is placed in multiple small tubes, alongside different dilutions of antibiotics.[11] Whether a bacteria is sensitive or not is determined by automatic optical methods, after a period of incubation.[6] Broth dilution is considered the gold standard for phenotypic testing.[11] The lowest concentration of antibiotics that inhibits growth is considered the MIC.[6]
Matrix-Assisted Laser Desorption Ionisation-Time of Flight Mass Spectrometry (MALDI-TOF MS) is another method of phenotypic susceptibility testing.[7] This is a form of time-of-flight mass spectrometry, in which the molecules of a bacterium are subject to matrix-assisted laser desorption.[13] The ionised particles are then accelerated, and spectral peaks recorded, producing an expression profile, which is capable of differentiating specific bacterial strains after being compared to known profiles.[13] This includes, in the context of antibiotic susceptibility testing, strains such as beta-lactamase producing E coli.[8] MALTI-TOF is rapid and automated.[8] There are limitations to testing in this format however; results may not match the results of phenotypic testing,[8] and acquisition and maintenance is expensive.[14]
Automated systems exist that replicate manual processes, for example, by using pictures and software analysis to report the zone of inhibition in diffusion testing. or dispensing samples and determining results in dilutional testing.[11]
Genetic methods
Genetic testing, such as via polymerase chain reaction, DNA microarray, DNA chips, and loop-mediated isothermal amplification, which may be used to detect whether bacteria possess genes which confer antibiotic resistance.[8][15] An example is the use of PCR to detect the mecA gene for beta-lactam resistant Staphylococcus aureus.[8] Other examples include assays for testing vancomycin resistance genes vanA and vanB in Enteroccocus species, and antibiotic resistance in Pseudomonas aeruginosa, Klebsiella pneumoniae and Escherichia coli.[8] These tests have the benefit of being direct and rapid, as compared with observable methods,[8] and have a high likelihood of detecting a finding when there is one to detect.[16] However, resistance genes detected do not always match whether a bacteria has resistance on a phenotypic method.[8] Unfortunately, the tests are also expensive and require specifically trained personnel.[14]
Polymerase chain reaction (PCR) is an method of identifying genes related to antibiotic susceptibility.[13] In the PCR process, the genome of a bacteria is denatured. Primers specific to a sought-after gene are added to a solution containing the DNA, and a DNA polymerase is added alongside a mixture containing molecules that will be needed (for example, nucleotides and ions).[14] If a gene is present, every time this process runs, the quantity of the target gene will be doubled.[14] After this process, the presence of the genes is demonstrated through a variety of methods including electrophoresis, southern blotting, and other DNA sequencing analysis methods.[14]
DNA microarrays and chips use the binding of complementary DNA to a target gene or nucleic acid sequence.[8] The benefit of this is that multiple genes can be assessed simultaneously.[8]
Reporting
The results of the testing are reported as a table, sometimes called an antibiogram.[17] Bacteria might be marked as sensitive, resistant, or having intermediate resistance to an antibiotic.[5] Specific patterns pf drug resistance or multi drug resistance may be noted, such as the presence of an extended-spectrum beta lactamase.[5]
The sensitive, resistant or intermediate resistance to antibiotics is reported based on the minimum inhibitory concentration. It is compared to known values for a given bacterium and antibiotic.[5] For example, Streptococcus pneumoniae isolates are considered susceptible to penicillin if MICs are ≤0.06 μg/ml, intermediate if MICs are 0.12 to 1 μg/ml, and resistant if MICs are ≥2 μg/ml.[18][19] Such information may be useful to the clinician, who can change the empirical treatment, to a more custom-tailored treatment that is directed only at the causative bacterium.[1][8] Sometimes, whether an antibiotic is marked as resistant is also based on bacterial characteristics that are associated with known methods of resistance such as the potential for beta lactamase production.[20][5]
Clinical practice
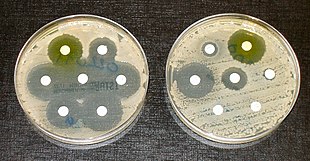
Ideal antibiotic therapy is based on determining the causal agent and its antibiotic sensitivity. Empirical treatment is often started before laboratory microbiological reports are available when treatment should not be delayed due to the seriousness of the disease. The effectiveness of individual antibiotics varies with the anatomical site of the infection, the ability of the antibiotic to reach the site of infection, and the ability of the bacteria to resist or inactivate the antibiotic.[22]
Often clinical specimens are sent to the clinical laboratory for culture and sensitivity, which is culture and antibiotic sensitivity testing offered as one combined service.[5] This is done through collecting samples from affected body sites, preferably before antibiotics are given. For example, a person in an intensive care unit may develop a hospital-acquired pneumonia. There is a chance the causal bacteria may be different to community-acquired pneumonia.[23] Treatment is generally started empirically, on the basis of surveillance data about the local common bacterial causes. This first treatment, based on statistical information about former patients, and aimed at a large group of potentially involved microbes, is called empirical treatment.[24]
Before starting this treatment, the physician will collect a sample from the site of a suspected infection: a blood culture sample when bacteria possibly have invaded the bloodstream, a sputum sample in the case of a ventilator associated pneumonia, and a urine sample in the case of a urinary tract infection. These samples are transferred to the microbiology laboratory where they are added to culture media.[25] Microscopy is not useful for all samples because often the normal bacterial flora cannot be distinguished from pathogens.[26] Additionally, when reported, a decision must be made for some bacteria such as staphylococcus epidermidis, as to whether they are the cause of an infection, or simply commensual bacteria or contaminants.[5][27]
When antibiotic sensitivity testing is reported, it will provide useful information about the organisms present in the sample, and which antibiotics will be effective.[5] Antibiotic sensitivity testing is done in a laboratory (in vitro), but the correlation of this testing to the sensitivity of the antibiotics in a person (in vivo) is often high enough for the test to be clinically useful.[28]
History
Since the discovery of the beta-lactam antibiotic penicillin, the rates of antimicrobial resistance have increased. Over time, methods for testing the sensitivity of bacteria to antibiotics have developed and changed.[14]
Alexander Fleming in the 1920s developed the first method of susceptibility testing. The "gutter method" that he developed was a diffusion method, involved an antibiotic that was diffused through a gutter made of agar.[14] In the 1940s, multiple investigators, including Pope, Foster and Woodruff, Vincent and Vincent used paper discs instead.[14] All these methods involve testing only susceptibility to penicillin.[14] The results were difficult to interpret and not reliable, because of inaccurate results that were not standardised between laboratories.[14]
Dilution has been used as a method to grow and identify bacteria since the 1870s, and as a method of testing the susceptibility of bacteria to antibiotics since 1929, also by Alexander Fleming.[14] The way of determining susceptibility changed from how turbid the solution was, to the pH (in 1942), to optical instruments.[14] The use of larger tube-based "macrodilution" testing has been superceded by smaller "microdilution" kits.[6]
In 1966, the World Health Organisation confirmed the Kirby-Bauer method as the standard method for susceptibility testing; it is simple, cost-effective and can test multiple antibiotics.[14]
The Etest was developed in 1980 by Bolmstrӧm and Eriksson, and MALDI-TOF developed in 2000s.[14] An array of automated systems has been developed since and after the 1980s.[14] PCR was the first genetic test available and first published as a method of detecting antibiotic susceptibility in 2001.[14]
Further research
Point-of-care testing is being developed to speed up the time for testing, and to help practitioners avoid prescribing unnecessary antibiotics in the style of precision medicine.[29] Traditional techniques typically take 12 to 48 hours,[7] to up to five days.[5] In contrast, rapid testing using molecular diagnostics is defined as "being feasible within an 8-h(our) working shift".[7] Progress has been slow due to a range of reasons including cost and regulation.[30]
Additional research is focused at the shortcomings of current testing methods. As well as the duration it takes to report of phenotypic methods, they are laborious, have difficult portability and are difficult to use in resource-limited settings, and have a chance of cross-contamination.[14]
As of 2017, point-of-care resistance diagnostics was available for methicillin-resistant Staphylococcus aureus (MRSA), rifampin-resistant Mycobacterium tuberculosis (TB), and Vancomycin-resistant enterococci (VRE) through GeneXpert by molecular diagnostics company Cepheid.[31]
Quantitative PCR, with the view of determining the percent of a detected bacteria that possesses a resistance gene, is being explored.[8] Whole genome sequencing of isolated bacteria is also being explored, and likely to become more available as costs decrease and speed increases over time.[8]
Additional methods explored include microfluidics, which uses tiny fluid and a variety of testing methods, such as optical, electrochemical, and magnetic.[8] Such assays do not require much fluid to be tested, are rapid and portable.[8]
The use of fluorescent dyes has been explored.[8] This are labelled proteins targeted at biomarkers, nucleic acid sequences present within cells that are found when the bacterium is resistant to an antibiotic.[8] An isolate of bacteria is fixed in position and then dissolved. The isolate is then exposed to fluoresent dye, which will be luminescent when viewed.[8]
Improvements to existing platforms are also being explored, including improvements in imaging systems that are able to more rapidly identify the MIC in phenotypic samples; or the use of bioluminescent enzymes that demonstrate bacterial growth to make changes more easily visible.[14]
See also
- Antibiotic resistance
- Miles-Misra method
- Bacterial culture
- Throat culture
- Laboratory specimen
Bibliography
References
- Leekha S, Terrell CL, Edson RS (February 2011). "General principles of antimicrobial therapy". Mayo Clinic Proceedings. 86 (2): 156–67. doi:10.4065/mcp.2010.0639. PMC 3031442. PMID 21282489.
Once microbiology results have helped to identify the etiologic pathogen and/or antimicrobial susceptibility data are available, every attempt should be made to narrow the antibiotic spectrum. This is a critically important component of antibiotic therapy because it can reduce cost and toxicity and prevent the emergence of antimicrobial resistance in the community
- Kang CI, Kim J, Park DW, Kim BN, Ha US, Lee SJ, et al. (March 2018). "Clinical Practice Guidelines for the Antibiotic Treatment of Community-Acquired Urinary Tract Infections". Infection & Chemotherapy. 50 (1): 67–100. doi:10.3947/ic.2018.50.1.67. PMC 5895837. PMID 29637759.
- Partridge SR, Kwong SM, Firth N, Jensen SO (October 2018). "Mobile Genetic Elements Associated with Antimicrobial Resistance". Clinical Microbiology Reviews. 31 (4). doi:10.1128/CMR.00088-17. PMC 6148190. PMID 30068738.
- Molton JS, Tambyah PA, Ang BS, Ling ML, Fisher DA (May 2013). Weinstein RA (ed.). "The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia". Clinical Infectious Diseases. 56 (9): 1310–8. doi:10.1093/cid/cit020. PMID 23334810.
- Giuliano C, Patel CR, Kale-Pradhan PB (April 2019). "A Guide to Bacterial Culture Identification And Results Interpretation". P & T. 44 (4): 192–200. PMC 6428495. PMID 30930604.
- Jorgensen JH, Ferraro MJ (December 2009). "Antimicrobial susceptibility testing: a review of general principles and contemporary practices". Clinical Infectious Diseases. 49 (11): 1749–55. doi:10.1086/647952. PMID 19857164.
- van Belkum A, Bachmann TT, Lüdke G, Lisby JG, Kahlmeter G, Mohess A, et al. (January 2019). "Developmental roadmap for antimicrobial susceptibility testing systems". Nature Reviews. Microbiology. 17 (1): 51–62. doi:10.1038/s41579-018-0098-9. PMC 7138758. PMID 30333569.
- Pulido MR, García-Quintanilla M, Martín-Peña R, Cisneros JM, McConnell MJ (December 2013). "Progress on the development of rapid methods for antimicrobial susceptibility testing". The Journal of Antimicrobial Chemotherapy. 68 (12): 2710–7. doi:10.1093/jac/dkt253. PMID 23818283.
- Syal K, Mo M, Yu H, Iriya R, Jing W, Guodong S, et al. (2017). "Current and emerging techniques for antibiotic susceptibility tests". Theranostics. 7 (7): 1795–1805. doi:10.7150/thno.19217. PMC 5479269. PMID 28638468.
- "Bauer-Kirby disk Diffusion". www.uphs.upenn.edu.
- Jorgensen, James H.; Turnidge, John D. (2015). "71. Susceptibility Test Methods: Dilution and Disk Diffusion Methods". Manual of Clinical Microbiology (11th ed.). pp. 1253–1273.
- Burnett 2005, p. 169.
- Bauer KA, Perez KK, Forrest GN, Goff DA (October 2014). "Review of rapid diagnostic tests used by antimicrobial stewardship programs". Clinical Infectious Diseases. 59 Suppl 3 (suppl_3): S134-45. doi:10.1093/cid/ciu547. PMID 25261540.
- Khan ZA, Siddiqui MF, Park S (May 2019). "Current and Emerging Methods of Antibiotic Susceptibility Testing". Diagnostics. 9 (2): 49. doi:10.3390/diagnostics9020049. PMC 6627445. PMID 31058811.
- Poirel L, Jayol A, Nordmann P (April 2017). "Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes". Clinical Microbiology Reviews. 30 (2): 557–596. doi:10.1128/CMR.00064-16. PMC 5355641. PMID 28275006.
- Arena, Fabio; Viaggi, Bruno; Galli, Luisa; Rossolini, Gian Maria (October 2015). "Antibiotic Susceptibility Testing: Present and Future". The Pediatric Infectious Disease Journal. 34 (10): 1128–1130. doi:10.1097/INF.0000000000000844.
- "Medical Definition of ANTIBIOGRAM". www.merriam-webster.com. Retrieved 2020-07-05.
- Jacobs MR, Bajaksouzian S, Palavecino-Fasola EL, Holoszyc HM, Appelbaum PC (January 1998). "Determination of penicillin MICs for Streptococcus pneumoniae by using a two- or three-disk diffusion procedure". Journal of Clinical Microbiology. 36 (1): 179–83. doi:10.1128/JCM.36.1.179-183.1998. PMC 124830. PMID 9431943.
- Goldsmith CE, Moore JE, Murphy PG (December 1997). "Pneumococcal resistance in the UK". The Journal of Antimicrobial Chemotherapy. 40 Suppl A: 11–8. doi:10.1093/jac/40.suppl_1.11. PMID 9484868.
- Winstanley T, Courvalin P (July 2011). "Expert systems in clinical microbiology". Clinical Microbiology Reviews. 24 (3): 515–56. doi:10.1128/CMR.00061-10. PMC 3131062. PMID 21734247.
- Kirby-Bauer Disk Diffusion Susceptibility Test Protocol Archived 26 June 2011 at the Wayback Machine, Jan Hudzicki, ASM
- Burnett 2005, p. 167.
- Peyrani P, Mandell L, Torres A, Tillotson GS (February 2019). "The burden of community-acquired bacterial pneumonia in the era of antibiotic resistance". Expert Review of Respiratory Medicine. 13 (2): 139–152. doi:10.1080/17476348.2019.1562339. PMID 30596308. S2CID 58640721.
- Burnett 2005, pp. 167–171.
- Burnett 2005, pp. 135–144.
- Pommerville 2018, p. 632.
- Becker K, Heilmann C, Peters G (October 2014). "Coagulase-negative staphylococci". Clinical Microbiology Reviews. 27 (4): 870–926. doi:10.1128/CMR.00109-13. PMC 4187637. PMID 25278577.
- Burnett 2005, p. 168.
- "Diagnostics Are Helping Counter Antimicrobial Resistance, But More Work Is Needed". MDDI Online. 2018-11-20. Retrieved 2018-12-02.
- "Progress on antibiotic resistance". Nature. 562 (7727): 307. October 2018. Bibcode:2018Natur.562Q.307.. doi:10.1038/d41586-018-07031-7. PMID 30333595.
- McAdams D (January 2017). "Resistance diagnosis and the changing epidemiology of antibiotic resistance". Annals of the New York Academy of Sciences. 1388 (1): 5–17. Bibcode:2017NYASA1388....5M. doi:10.1111/nyas.13300. PMID 28134444.