Amphiphysin
Amphiphysin is a protein that in humans is encoded by the AMPH gene.[5][6]
Function
This gene encodes a protein associated with the cytoplasmic surface of synaptic vesicles. A subset of patients with stiff person syndrome who were also affected by breast cancer are positive for autoantibodies against this protein. Alternate splicing of this gene results in two transcript variants encoding different isoforms. Additional splice variants have been described, but their full length sequences have not been determined.[6]
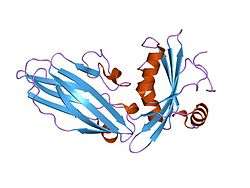
Amphiphysin is a brain-enriched protein with an N-terminal lipid interaction, dimerisation and membrane bending BAR domain, a middle clathrin and adaptor binding domain and a C-terminal SH3 domain. In the brain, its primary function is thought to be the recruitment of dynamin to sites of clathrin-mediated endocytosis. There are 2 mammalian amphiphysins with similar overall structure. A ubiquitous splice form of amphiphysin-2 (BIN1) that does not contain clathrin or adaptor interactions is highly expressed in muscle tissue and is involved in the formation and stabilization of the T-tubule network. In other tissues amphiphysin is likely involved in other membrane bending and curvature stabilization events.
Interactions
Amphiphysin has been shown to interact with DNM1,[7][8][9][10][11] Phospholipase D1,[12] CDK5R1,[13] PLD2,[12] CABIN1[14] and SH3GL2.[7][15]
References
- GRCh38: Ensembl release 89: ENSG00000078053 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000021314 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- De Camilli P, Thomas A, Cofiell R, Folli F, Lichte B, Piccolo G, Meinck HM, Austoni M, et al. (December 1993). "The synaptic vesicle-associated protein amphiphysin is the 128-kD autoantigen of Stiff-Man syndrome with breast cancer". The Journal of Experimental Medicine. 178 (6): 2219–23. doi:10.1084/jem.178.6.2219. PMC 2191289. PMID 8245793.
- "Entrez Gene: AMPH amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen)".
- Micheva KD, Kay BK, McPherson PS (October 1997). "Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin". The Journal of Biological Chemistry. 272 (43): 27239–45. doi:10.1074/jbc.272.43.27239. PMID 9341169.
- Wigge P, Köhler K, Vallis Y, Doyle CA, Owen D, Hunt SP, McMahon HT (October 1997). "Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis". Molecular Biology of the Cell. 8 (10): 2003–15. doi:10.1091/mbc.8.10.2003. PMC 25662. PMID 9348539.
- McMahon HT, Wigge P, Smith C (August 1997). "Clathrin interacts specifically with amphiphysin and is displaced by dynamin". FEBS Letters. 413 (2): 319–22. doi:10.1016/S0014-5793(97)00928-9. PMID 9280305.
- Chen-Hwang MC, Chen HR, Elzinga M, Hwang YW (May 2002). "Dynamin is a minibrain kinase/dual specificity Yak1-related kinase 1A substrate". The Journal of Biological Chemistry. 277 (20): 17597–604. doi:10.1074/jbc.M111101200. PMID 11877424.
- Grabs D, Slepnev VI, Songyang Z, David C, Lynch M, Cantley LC, De Camilli P (May 1997). "The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence". The Journal of Biological Chemistry. 272 (20): 13419–25. doi:10.1074/jbc.272.20.13419. PMID 9148966.
- Lee C, Kim SR, Chung JK, Frohman MA, Kilimann MW, Rhee SG (June 2000). "Inhibition of phospholipase D by amphiphysins". The Journal of Biological Chemistry. 275 (25): 18751–8. doi:10.1074/jbc.M001695200. PMID 10764771.
- Floyd SR, Porro EB, Slepnev VI, Ochoa GC, Tsai LH, De Camilli P (March 2001). "Amphiphysin 1 binds the cyclin-dependent kinase (cdk) 5 regulatory subunit p35 and is phosphorylated by cdk5 and cdc2". The Journal of Biological Chemistry. 276 (11): 8104–10. doi:10.1074/jbc.M008932200. PMID 11113134.
- Lai MM, Luo HR, Burnett PE, Hong JJ, Snyder SH (November 2000). "The calcineurin-binding protein cain is a negative regulator of synaptic vesicle endocytosis". The Journal of Biological Chemistry. 275 (44): 34017–20. doi:10.1074/jbc.C000429200. PMID 10931822.
- Modregger J, Schmidt AA, Ritter B, Huttner WB, Plomann M (February 2003). "Characterization of Endophilin B1b, a brain-specific membrane-associated lysophosphatidic acid acyl transferase with properties distinct from endophilin A1". The Journal of Biological Chemistry. 278 (6): 4160–7. doi:10.1074/jbc.M208568200. PMID 12456676.
Further reading
- Lichte B, Veh RW, Meyer HE, Kilimann MW (July 1992). "Amphiphysin, a novel protein associated with synaptic vesicles". The EMBO Journal. 11 (7): 2521–30. doi:10.1002/j.1460-2075.1992.tb05317.x. PMC 556727. PMID 1628617.
- Yamamoto R, Li X, Winter S, Francke U, Kilimann MW (February 1995). "Primary structure of human amphiphysin, the dominant autoantigen of paraneoplastic stiff-man syndrome, and mapping of its gene (AMPH) to chromosome 7p13-p14". Human Molecular Genetics. 4 (2): 265–8. doi:10.1093/hmg/4.2.265. PMID 7757077.
- David C, Solimena M, De Camilli P (August 1994). "Autoimmunity in stiff-Man syndrome with breast cancer is targeted to the C-terminal region of human amphiphysin, a protein similar to the yeast proteins, Rvs167 and Rvs161". FEBS Letters. 351 (1): 73–9. doi:10.1016/0014-5793(94)00826-4. PMID 8076697.
- David C, McPherson PS, Mundigl O, de Camilli P (January 1996). "A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals". Proceedings of the National Academy of Sciences of the United States of America. 93 (1): 331–5. doi:10.1073/pnas.93.1.331. PMC 40232. PMID 8552632.
- Grabs D, Slepnev VI, Songyang Z, David C, Lynch M, Cantley LC, De Camilli P (May 1997). "The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence". The Journal of Biological Chemistry. 272 (20): 13419–25. doi:10.1074/jbc.272.20.13419. PMID 9148966.
- Butler MH, David C, Ochoa GC, Freyberg Z, Daniell L, Grabs D, Cremona O, De Camilli P (June 1997). "Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle". The Journal of Cell Biology. 137 (6): 1355–67. doi:10.1083/jcb.137.6.1355. PMC 2132527. PMID 9182667.
- Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS (June 1997). "Identification and characterization of a nerve terminal-enriched amphiphysin isoform". The Journal of Biological Chemistry. 272 (26): 16700–6. doi:10.1074/jbc.272.26.16700. PMID 9195986.
- McMahon HT, Wigge P, Smith C (August 1997). "Clathrin interacts specifically with amphiphysin and is displaced by dynamin". FEBS Letters. 413 (2): 319–22. doi:10.1016/S0014-5793(97)00928-9. PMID 9280305.
- Micheva KD, Ramjaun AR, Kay BK, McPherson PS (September 1997). "SH3 domain-dependent interactions of endophilin with amphiphysin". FEBS Letters. 414 (2): 308–12. doi:10.1016/S0014-5793(97)01016-8. PMID 9315708.
- Wigge P, Köhler K, Vallis Y, Doyle CA, Owen D, Hunt SP, McMahon HT (October 1997). "Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis". Molecular Biology of the Cell. 8 (10): 2003–15. doi:10.1091/mbc.8.10.2003. PMC 25662. PMID 9348539.
- Floyd S, Butler MH, Cremona O, David C, Freyberg Z, Zhang X, Solimena M, Tokunaga A, et al. (January 1998). "Expression of amphiphysin I, an autoantigen of paraneoplastic neurological syndromes, in breast cancer". Molecular Medicine. 4 (1): 29–39. doi:10.1007/BF03401727. PMC 2230265. PMID 9513187.
- Ramjaun AR, McPherson PS (June 1998). "Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites". Journal of Neurochemistry. 70 (6): 2369–76. doi:10.1046/j.1471-4159.1998.70062369.x. PMID 9603201.
- Slepnev VI, Ochoa GC, Butler MH, Grabs D, De Camilli P (August 1998). "Role of phosphorylation in regulation of the assembly of endocytic coat complexes". Science. 281 (5378): 821–4. doi:10.1126/science.281.5378.821. PMID 9694653.
- Cestra G, Castagnoli L, Dente L, Minenkova O, Petrelli A, Migone N, Hoffmüller U, Schneider-Mergener J, Cesareni G (November 1999). "The SH3 domains of endophilin and amphiphysin bind to the proline-rich region of synaptojanin 1 at distinct sites that display an unconventional binding specificity". The Journal of Biological Chemistry. 274 (45): 32001–7. doi:10.1074/jbc.274.45.32001. PMID 10542231.
- Slepnev VI, Ochoa GC, Butler MH, De Camilli P (June 2000). "Tandem arrangement of the clathrin and AP-2 binding domains in amphiphysin 1 and disruption of clathrin coat function by amphiphysin fragments comprising these sites". The Journal of Biological Chemistry. 275 (23): 17583–9. doi:10.1074/jbc.M910430199. PMID 10748223.
- Lee C, Kim SR, Chung JK, Frohman MA, Kilimann MW, Rhee SG (June 2000). "Inhibition of phospholipase D by amphiphysins". The Journal of Biological Chemistry. 275 (25): 18751–8. doi:10.1074/jbc.M001695200. PMID 10764771.
- Onofri F, Giovedi S, Kao HT, Valtorta F, Bongiorno Borbone L, De Camilli P, Greengard P, Benfenati F (September 2000). "Specificity of the binding of synapsin I to Src homology 3 domains". The Journal of Biological Chemistry. 275 (38): 29857–67. doi:10.1074/jbc.M006018200. PMID 10899172.
- Lai MM, Luo HR, Burnett PE, Hong JJ, Snyder SH (November 2000). "The calcineurin-binding protein cain is a negative regulator of synaptic vesicle endocytosis". The Journal of Biological Chemistry. 275 (44): 34017–20. doi:10.1074/jbc.C000429200. PMID 10931822.
- Floyd SR, Porro EB, Slepnev VI, Ochoa GC, Tsai LH, De Camilli P (March 2001). "Amphiphysin 1 binds the cyclin-dependent kinase (cdk) 5 regulatory subunit p35 and is phosphorylated by cdk5 and cdc2". The Journal of Biological Chemistry. 276 (11): 8104–10. doi:10.1074/jbc.M008932200. PMID 11113134.
- Leventis PA, Chow BM, Stewart BA, Iyengar B, Campos AR, Boulianne GL (November 2001). "Drosophila Amphiphysin is a post-synaptic protein required for normal locomotion but not endocytosis". Traffic. 2 (11): 839–50. doi:10.1034/j.1600-0854.2001.21113.x. PMID 11733051.
- Zhang B, Zelhof AC (July 2002). "Amphiphysins: raising the BAR for synaptic vesicle recycling and membrane dynamics. Bin-Amphiphysin-Rvsp". Traffic. 3 (7): 452–60. doi:10.1034/j.1600-0854.2002.30702.x. PMID 12047553. Review.
- Zelhof AC, Bao H, Hardy RW, Razzaq A, Zhang B, Doe CQ (December 2001). "Drosophila Amphiphysin is implicated in protein localization and membrane morphogenesis but not in synaptic vesicle endocytosis". Development. 128 (24): 5005–15. PMID 11748137.
- Mathew D, Popescu A, Budnik V (November 2003). "Drosophila amphiphysin functions during synaptic Fasciclin II membrane cycling". The Journal of Neuroscience. 23 (33): 10710–6. doi:10.1523/JNEUROSCI.23-33-10710.2003. PMID 14627656.
- Takei K, Slepnev VI, Haucke V, De Camilli P (May 1999). "Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis". Nature Cell Biology. 1 (1): 33–9. doi:10.1038/9004. PMID 10559861.
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (January 2004). "BAR domains as sensors of membrane curvature: the amphiphysin BAR structure". Science. 303 (5657): 495–9. doi:10.1126/science.1092586. PMID 14645856.
- Evergren E, Marcucci M, Tomilin N, Löw P, Slepnev V, Andersson F, Gad H, Brodin L, et al. (July 2004). "Amphiphysin is a component of clathrin coats formed during synaptic vesicle recycling at the lamprey giant synapse". Traffic. 5 (7): 514–28. doi:10.1111/j.1398-9219.2004.00198.x. PMID 15180828.
- Razzaq A, Robinson IM, McMahon HT, Skepper JN, Su Y, Zelhof AC, Jackson AP, Gay NJ, O'Kane CJ (November 2001). "Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila". Genes & Development. 15 (22): 2967–79. doi:10.1101/gad.207801. PMC 312829. PMID 11711432.
External links
- Bringing your curves to the bar, amphiphysin home page
- Human AMPH genome location and AMPH gene details page in the UCSC Genome Browser.