6S / SsrS RNA
In the field of molecular biology the 6S RNA is a noncoding RNA that was one of the first to be identified and sequenced.[1] What it does in the bacterial cell was unknown until recently. In the early 2000s scientists found out the function of 6S RNA to as a regulator of sigma 70-dependent gene transcription. All bacterial RNA polymerases have a subunit called a sigma factor. The sigma factors are important because they control how DNA promoter binding and RNA transcription start sites. Sigma 70 was the first one to be discovered in Escherichia coli.[2][3]
| 6S / SsrS RNA | |
|---|---|
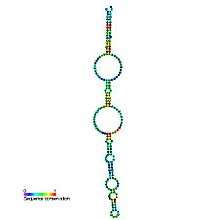 Predicted secondary structure and sequence conservation of 6S | |
| Identifiers | |
| Symbol | 6S |
| Rfam | RF00013 |
| Other data | |
| RNA type | Gene |
| Domain(s) | Bacteria |
| SO | 0000376 |
| PDB structures | PDBe |
Structure
The structure of 6S RNA was defined in 1971.[2] It is a small RNA strand consisting of 184 nucleotides. 6S RNA is a long double-stranded structure and has a single strand loop. Its structure is similar to an open promoter complex of DNA structure. Varies analyses discovered that 6S RNAs are capable of forming a secondary structure. The secondary structure consists of two irregular helical stem regions, making a large core loop which is called a central knot.
Function and Regulation
The function of 6S RNA is to regulate transcription for E. Coli cell survival because it is essential in the process.[4] 6S RNA specifically associates with RNA polymerase holoenzyme containing the sigma70 specificity factor. This interaction represses expression from sigma70-dependent promoters during stationary phase.[5] Which will lead to activate the transcription from sigma 70 dependent promoters. Therefore, during the change in E. coli from logarithmic growth to stationary phase, the 6S RNA performs as a regulator of transcription. S RNA homologs have recently been identified in most bacterial genomes.[3][6] polymerase holoenzyme, which contains the housekeeping sigma factor and be expressed during different stages of growth. In many proteobacteria, 6S RNA may be processed from a transcript encoding homologs of the E. coli YgfA protein, which is a putative methenyltetrahydrofolate synthetase. Diverged 6S RNAs have been identified in additional bacterial lineages.[7][8] The purD RNA motif has been experimentally shown to overlap with 6S RNA.[7] One way to examine if the activity of 6S RNA by doing a knockout of 6S RNA. Strains with mutations in 6S RNA have a reduction of lifespan in contrast to the wild-type cells after more than 20 days of nonstop culture. When mutant 6S cells cultured with wild-type cells, it will be at a modest disadvantage in the following days of growth.[9]
References
- Brownlee GG (February 1971). "Sequence of 6S RNA of E. coli". Nature. 229 (5): 147–149. doi:10.1038/229147a0. PMID 4929322.
- Lee JY, Park H, Bak G, Kim KS, Lee Y (September 2013). "Regulation of transcription from two ssrS promoters in 6S RNA biogenesis". Molecules and Cells. 36 (3): 227–234. doi:10.1007/s10059-013-0082-1. PMC 3887979. PMID 23864284.
- Trotochaud AE, Wassarman KM (April 2005). "A highly conserved 6S RNA structure is required for regulation of transcription". Nature Structural & Molecular Biology. 12 (4): 313–319. doi:10.1038/nsmb917. PMID 15793584.
- Steuten B, Hoch PG, Damm K, Schneider S, Köhler K, Wagner R, Hartmann RK (2014-05-01). "Regulation of transcription by 6S RNAs: insights from the Escherichia coli and Bacillus subtilis model systems". RNA Biology. 11 (5): 508–521. doi:10.4161/rna.28827. PMC 4152359. PMID 24786589.
- Wassarman KM, Storz G (June 2000). "6S RNA regulates E. coli RNA polymerase activity". Cell. 101 (6): 613–623. doi:10.1016/S0092-8674(00)80873-9. PMID 10892648.
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR (May 2005). "6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter". RNA. 11 (5): 774–784. doi:10.1261/rna.7286705. PMC 1370762. PMID 15811922.
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. (March 2010). "The primary transcriptome of the major human pathogen Helicobacter pylori". Nature. 464 (7286): 250–255. Bibcode:2010Natur.464..250S. doi:10.1038/nature08756. PMID 20164839.
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes". Genome Biology. 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571. PMID 20230605.
- Steuten B, Hoch PG, Damm K, Schneider S, Köhler K, Wagner R, Hartmann RK (2014-05-01). "Regulation of transcription by 6S RNAs: insights from the Escherichia coli and Bacillus subtilis model systems". RNA Biology. 11 (5): 508–521. doi:10.4161/rna.28827. PMC 4152359. PMID 24786589.