6-O-Methylguanine
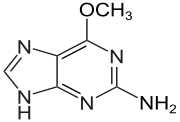
6-O-Methylguanine is a derivative of the nucleobase guanine in which a methyl group is attached to the oxygen atom. It base-pairs to thymine rather than cytosine, causing a G:C to A:T transition in DNA.
 | |
| Names | |
|---|---|
| IUPAC name
6-Methoxy-7H-purin-2-amine | |
| Other names
6-Methoxyguanine; O6-Methylguanine; 2-Amino-6-methoxypurine | |
| Identifiers | |
3D model (JSmol) |
|
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.111.933 |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C6H7N5O | |
| Molar mass | 165.156 g·mol−1 |
| Melting point | >300 °C |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Formation
6-O-Methylguanine is formed in DNA by alkylation of the oxygen atom of guanine, most often by N-nitroso compounds (NOC)[2] and sometimes due to methylation by other compounds such as endogenous S-adenosyl methionine.[3] NOC are alkylating agents formed by the reaction of nitrite or other nitrogen oxides with secondary amines and N-alkylamides, yielding N-alkylnitrosamines and N-alkylnitrosamides.[2]
NOC are found in some foods (bacon, sausages, cheese) and tobacco smoke, and are formed in the gastrointestinal tract, especially after consumption of red meat.[2] In addition, endogenous nitric oxide levels were found to be enhanced under chronic inflammatory conditions, and this could favor NOC formation in the large intestine.
Repair and carcinogenicity
Repair of 6-O-methylguanine in DNA is primarily carried out by O-6-methylguanine-DNA methyltransferase (MGMT). Epigenetic reductions in MGMT expression are one of the most frequent DNA repair defects, associated with carcinogenesis.[4] (Also see MGMT expression in cancer.)
Mutagenicity
In 1985 Yarosh summarized the early work that established 6-O-methylguanine as the alkylated base in DNA that was the most mutagenic and carcinogenic.[5] In 1994 Rasouli-Nia et al.[6] showed that about one mutation was induced for every eight unrepaired 6-O-Methylguanines in DNA.
About one third of the time 6-O-methylguanine mispairs during replication, leading to the incorporation of dTMP rather than dCMP.[7] 6-O-methylguanine is therefore a mutagenic nucleobase. However, the mutagenicity of a particular 6-O-methylguanine base depends on the sequence in which it is embedded.[8]
Other effects
Unrepaired 6-O-methylguanine can also lead to cell cycle arrest, sister chromatid exchange, or apoptosis.[9] These effects are due to interaction of the DNA mismatch repair pathway with 6-O-methylguanine, and also depend on signaling network activation, led by early ATM, H2AX, CHK1 and p53 phosphorylation.[9]
References
- 6-O-Methylguanine at Sigma-Aldrich
- Fahrer J, Kaina B (2013). "O6-methylguanine-DNA methyltransferase in the defense against N-nitroso compounds and colorectal cancer". Carcinogenesis. 34 (11): 2435–42. doi:10.1093/carcin/bgt275. PMID 23929436.
- De Bont R, van Larebeke N (2004). "Endogenous DNA damage in humans: a review of quantitative data". Mutagenesis. 19 (3): 169–85. doi:10.1093/mutage/geh025. PMID 15123782.
- Bernstein C, Bernstein H (2015). "Epigenetic reduction of DNA repair in progression to gastrointestinal cancer". World J Gastrointest Oncol. 7 (5): 30–46. doi:10.4251/wjgo.v7.i5.30. PMC 4434036. PMID 25987950.
- Yarosh DB (1985). "The role of O6-methylguanine-DNA methyltransferase in cell survival, mutagenesis and carcinogenesis". Mutat. Res. 145 (1–2): 1–16. doi:10.1016/0167-8817(85)90034-3. PMID 3883145.
- Rasouli-Nia A, Sibghat-Ullah, Mirzayans R, Paterson MC, Day RS (1994). "On the quantitative relationship between O6-methylguanine residues in genomic DNA and production of sister-chromatid exchanges, mutations and lethal events in a Mer- human tumor cell line". Mutat. Res. 314 (2): 99–113. doi:10.1016/0921-8777(94)90074-4. PMID 7510369.
- Abbott PJ, Saffhill R (1979). "DNA synthesis with methylated poly(dC-dG) templates. Evidence for a competitive nature to miscoding by O(6)-methylguanine". Biochim. Biophys. Acta. 562 (1): 51–61. doi:10.1016/0005-2787(79)90125-4. PMID 373805.
- Georgiadis P, Smith CA, Swann PF (1991). "Nitrosamine-induced cancer: selective repair and conformational differences between O6-methylguanine residues in different positions in and around codon 12 of rat H-ras". Cancer Res. 51 (21): 5843–50. PMID 1933853.
- Noonan EM, Shah D, Yaffe MB, Lauffenburger DA, Samson LD (2012). "O6-Methylguanine DNA lesions induce an intra-S-phase arrest from which cells exit into apoptosis governed by early and late multi-pathway signaling network activation". Integrative Biology. 4 (10): 1237–55. doi:10.1039/c2ib20091k. PMC 3574819. PMID 22892544.