Directionality (molecular biology)
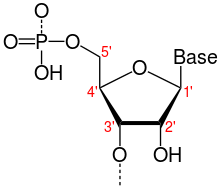
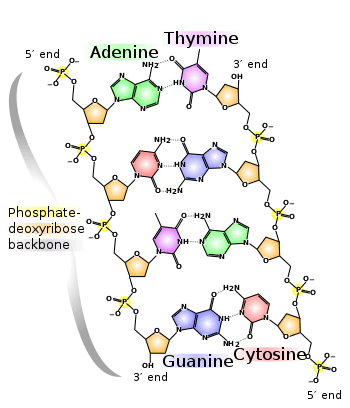
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide sugar-ring means that there will be a 5′-end (usually pronounced "five prime end" ), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′-end (usually pronounced "three prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information.
Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleoside monophosphates to the 3′-hydroxyl (-OH) group, via a phosphodiester bond. The relative positions of structures along a strand of nucleic acid, including genes and various protein binding sites, are usually noted as being either upstream (towards the 5′-end) or downstream (towards the 3′-end). (See also upstream and downstream.)
Directionality is related to, but independent from, sense. Transcription of single-stranded RNA from a double-stranded DNA template requires the selection of one strand of the DNA template as the template strand that directly interacts with the nascent RNA due to complementary sequence. The other strand is not copied directly, but necessarily its sequence will be similar to that of the RNA. Transcription initiation sites generally occur on both strands of an organism's DNA, and specify the location, direction, and circumstances under which transcription will occur. If the transcript encodes one or (rarely) more proteins, translation of each protein by the ribosome will proceed in a 5′ to 3′ direction, and will extend the protein from its N terminus toward its C terminus. For example, in a typical gene a start codon (5′-ATG-3′) is a DNA sequence within the sense strand. Transcription begins at an upstream site (relative to the sense strand), and as it proceeds through the region it copies the 3′-TAC-5′ from the template strand to produce 5′-AUG-3′ within a messenger RNA (mRNA). The mRNA is scanned by the ribosome from the 5′ end, where the start codon directs the incorporation of a methionine (prokaryotes use N-Formylmethionine instead) at the N terminus of the protein. By convention, single strands of DNA and RNA sequences are written in a 5′-to-3′ direction except as needed to illustrate the pattern of base pairing.
5′-end
The 5′-end (pronounced "five prime end") designates the end of the DNA or RNA strand that has the fifth carbon in the sugar-ring of the deoxyribose or ribose at its terminus. A phosphate group attached to the 5′-end permits ligation of two nucleotides, i.e., the covalent binding of a 5′-phosphate to the 3′-hydroxyl group of another nucleotide, to form a phosphodiester bond. Removal of the 5′-phosphate prevents ligation. To prevent unwanted nucleic acid ligation (e.g. self-ligation of a plasmid vector in DNA cloning), molecular biologists commonly remove the 5′-phosphate with a phosphatase.
The 5′-end of nascent messenger RNA is the site at which post-transcriptional capping occurs, a process which is vital to producing mature messenger RNA. Capping increases the stability of the messenger RNA while it undergoes translation, providing resistance to the degradative effects of exonucleases. It consists of a methylated nucleotide (methylguanosine) attached to the messenger RNA in a rare 5′- to 5′-triphosphate linkage.
The 5′-flanking region of a gene often denotes a region of DNA which is not transcribed into RNA. The 5′-flanking region contains the gene promoter, and may also contain enhancers or other protein binding sites.
The 5′-untranslated region (5′-UTR) is a region of a gene which is transcribed into mRNA, and is located at the 5′-end of the mRNA. This region of an mRNA may or may not be translated, but is usually involved in the regulation of translation. The 5′-untranslated region is the portion of the DNA starting from the cap site and extending to the base just before the AUG translation initiation codon of the main coding sequence. This region may have sequences, such as the ribosome binding site and Kozak sequence, which determine the translation efficiency of the mRNA, or which may affect the stability of the mRNA.
3′-end
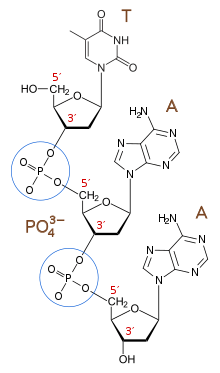
The 3′-end (three prime end) of a strand is so named due to it terminating at the hydroxyl group of the third carbon in the sugar-ring, and is known as the tail end. The 3′-hydroxyl is necessary in the synthesis of new nucleic acid molecules as it is ligated (joined) to the 5′-phosphate of a separate nucleotide, allowing the formation of strands of linked nucleotides.
Molecular biologists can use nucleotides that lack a 3′-hydroxyl (dideoxyribonucleotides) to interrupt the replication of DNA. This technique is known as the dideoxy chain-termination method or the Sanger method, and is used to determine the order of nucleotides in DNA.
The 3′-end of nascent messenger RNA is the site of post-transcriptional polyadenylation, which attaches a chain of 50 to 250 adenosine residues to produce mature messenger RNA. This chain helps in determining how long the messenger RNA lasts in the cell, influencing how much protein is produced from it.
The 3′-flanking region is a region of DNA that is not copied into the mature mRNA, but which is present adjacent to 3′-end of the gene. It was originally thought that the 3′-flanking DNA was not transcribed at all, but it was discovered to be transcribed into RNA and quickly removed during processing of the primary transcript to form the mature mRNA. The 3′-flanking region often contains sequences that affect the formation of the 3′-end of the message. It may also contain enhancers or other sites to which proteins may bind.
The 3′-untranslated region (3′-UTR) is a region of the DNA which is transcribed into mRNA and becomes the 3′-end of the message, but which does not contain protein coding sequence. Everything between the stop codon and the polyA tail is considered to be 3′-untranslated. The 3′-untranslated region may affect the translation efficiency of the mRNA or the stability of the mRNA. It also has sequences which are required for the addition of the poly(A) tail to the message, including the hexanucleotide AAUAAA.
See also
Further reading
- Harvey Lodish; Arnold Berk; Paul Matsudaira; Chris A. Kaiser (2004). Molecular Cell Biology (5th ed.). New York City: W.H. Freeman and Company. ISBN 978-0-7167-4366-8.