DAPI
DAPI (pronounced 'DAPPY'), or 4′,6-diamidino-2-phenylindole, is a fluorescent stain that binds strongly to adenine–thymine-rich regions in DNA. It is used extensively in fluorescence microscopy. As DAPI can pass through an intact cell membrane, it can be used to stain both live and fixed cells, though it passes through the membrane less efficiently in live cells and therefore provides a marker for membrane viability.
 | |
 | |
| Names | |
|---|---|
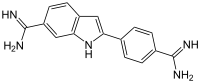
| IUPAC name
2-(4-Amidinophenyl)-1H-indole-6-carboxamidine | |
| Other names
4′,6-Diamidino-2-phenylindole | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
PubChem CID |
|
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C16H15N5 | |
| Molar mass | 277.331 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
History
DAPI was first synthesised in 1971 in the laboratory of Otto Dann as part of a search for drugs to treat trypanosomiasis. Although it was unsuccessful as a drug, further investigation indicated it bound strongly to DNA and became more fluorescent when bound. This led to its use in identifying mitochondrial DNA in ultracentrifugation in 1975, the first recorded use of DAPI as a fluorescent DNA stain.[1]
Strong fluorescence when bound to DNA led to the rapid adoption of DAPI for fluorescent staining of DNA for fluorescence microscopy. Its use for detecting DNA in plant, metazoa and bacteria cells and virus particles was demonstrated in the late 1970s, and quantitative staining of DNA inside cells was demonstrated in 1977. Use of DAPI as a DNA stain for flow cytometry was also demonstrated around this time.[1]
Fluorescence properties
When bound to double-stranded DNA, DAPI has an absorption maximum at a wavelength of 358 nm (ultraviolet) and its emission maximum is at 461 nm (blue). Therefore, for fluorescence microscopy, DAPI is excited with ultraviolet light and is detected through a blue/cyan filter. The emission peak is fairly broad.[2] DAPI will also bind to RNA, though it is not as strongly fluorescent. Its emission shifts to around 500 nm when bound to RNA.[3][4]
DAPI's blue emission is convenient for microscopists who wish to use multiple fluorescent stains in a single sample. There is some fluorescence overlap between DAPI and green-fluorescent molecules like fluorescein and green fluorescent protein (GFP) but the effect of this is small. Use of spectral unmixing can account for this effect if extremely precise image analysis is required.
Outside of analytical fluorescence light microscopy DAPI is also popular for labeling of cell cultures to detect the DNA of contaminating Mycoplasma or virus. The labelled Mycoplasma or virus particles in the growth medium fluoresce once stained by DAPI making them easy to detect.[5]
Modelling of absorption and fluorescence properties
This DNA fluorescent probe has been effectively modeled[6] using the time-dependent density functional theory, coupled with the IEF version of the polarizable continuum model. This quantum-mechanical modeling has rationalized the absorption and fluorescence behavior given by minor groove binding and intercalation in the DNA pocket, in term of a reduced structural flexibility and polarization.
Live cells and toxicity
DAPI can be used for fixed cell staining. The concentration of DAPI needed for live cell staining is generally very high; it is rarely used for live cells.[7] It is labeled non-toxic in its MSDS[8] and though it was not shown to have mutagenicity to E. coli,[9] it is labelled as a known mutagen in manufacturer information.[2] As it is a DNA binding compound, it is likely to have some low level mutagenic properties and care should be taken in its handling and disposal.
Alternatives
The Hoechst stains are similar to DAPI in that they are also blue-fluorescent DNA stains which are compatible with both live- and fixed-cell applications, as well as visible using the same equipment filter settings as for DAPI.
Notes and references
- Kapuscinski, J. (September 1995). "DAPI: a DNA-specific fluorescent probe". Biotech. Histochem. 70 (5): 220–233. doi:10.3109/10520299509108199. PMID 8580206.
- Invitrogen, DAPI Nucleic Acid Stain Archived 2009-03-06 at the Wayback Machine. accessed 2009-12-08.
- Scott Prahl, DAPI. accessed 2009-12-08.
- Kapuscinski J., . accessed 2013-02-25.
- RUSSELL, W. C.; NEWMAN, CAROL; WILLIAMSON, D. H. (1975). "A simple cytochemical technique for demonstration of DNA in cells infected with mycoplasmas and viruses". Nature. 253 (5491): 461–462. Bibcode:1975Natur.253..461R. doi:10.1038/253461a0. PMID 46112.
- A. Biancardi; T. Biver; F. Secco; B. Mennucci (2013). "An investigation of the photophysical properties of minor groove bound and intercalated DAPI through quantum-mechanical and spectroscopic tools". Phys. Chem. Chem. Phys. 15 (13): 4596–603. Bibcode:2013PCCP...15.4596B. doi:10.1039/C3CP44058C. PMID 23423468.
- Zink D, Sadoni N, Stelzer E (2003). "Visualizing Chromatin and Chromosomes in Living Cells. Usually for the live cells staining Hoechst Staining is used. DAPI gives a higher signal in the fixed cells compare to Hoechst Stain but in the live cells Hoechst Stain is used". Methods. 29 (1): 42–50. doi:10.1016/S1046-2023(02)00289-X. PMID 12543070.
- http://www.kpl.com/docs/msds/710301.pdf
- Ohta T, Tokishita S, Yamagata H (2001). "Ethidium bromide and SYBR Green I enhance the genotoxicity of UV-irradiation and chemical mutagens in E. coli". Mutat. Res. 492 (1–2): 91–7. doi:10.1016/S1383-5718(01)00155-3. PMID 11377248.
See also
| Wikimedia Commons has media related to DAPI. |
- DNA binding ligand
- Hoechst stain
- Lexitropsin
- Netropsin
- Pentamidine
