23S ribosomal RNA
The 23S rRNA is a 2904 nt long (in E. coli) component of the large subunit (50S) of the bacterial/archean ribosome. The ribosomal peptidyl transferase activity resides in domain V of this rRNA, and this domain is the most common binding site for antibiotics that inhibit translation. A well-known member of this antibiotic class, chloramphenicol, acts by inhibiting peptide bond formation, with recent 3D-structural studies showing two different binding sites depending on the species of ribosome. Linezolid and quinupristin-dalfopristin also bind to the 23S rRNA, and cross-resistance has been demonstrated between these antibiotics. Compared to 16S rRNA genes, 23S rRNA genes typically have higher sequence variations including insertions and/or deletions.[2]
| Pseudoknot of the domain G(G12) of 23S ribosomal RNA | |
|---|---|
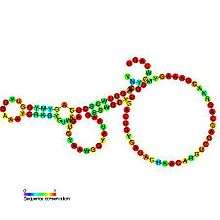 Predicted secondary structure and sequence conservation of PK-G12rRNA | |
| Identifiers | |
| Symbol | PK-G12rRNA |
| Rfam | RF01118 |
| Other data | |
| RNA type | Gene; rRNA |
| Domain(s) | Bacteria |
| SO | 0001263 |
| PDB structures | PDBe |
The eukaryotic homolog of the 23S rRNA is the 28S ribosomal RNA, with a region filled by the 5.8S ribosomal RNA.[3]
23S rRNA Functions
In general, rRNA has an essential function of peptidyl transferase, the stimulating core of the ribosome plays role in the peptide bond configuration. Both peptidyl-tRNA and aminoacyl-tRNA are important for synthesizing protein and transpeptidation response. However, 23S rRNA positions which are G2252, A2451, U2506, and U2585 have a significant function for tRNA binding in P site of the large ribosomal subunit. These modification nucleotides in site P can inhibit peptidyl-tRNA from binding and also remains U2555 modification intervene with transferring peptidyl-tRNA to puromycin. Furthermore, the chemical modification of half of these positions G2251, G2253, A2439, and U2584 can not prevent the tRNA binding. Peptidyl-tRNA of 50s subunits which binds to the P site preserve eight positions of 23S rRNA from chemical modification. On the other hand, 23S rRNA impacts on mutation for cell growth. Mutations A1912G, A1919G and Ψ1917C have a powerful growth phenotypes and they prevent translation while mutation A1916G has a simple growth phenotype and it leads to defect in the 50S subunits.[4]
See also
References
- Mueller F, Sommer I, Baranov P, Matadeen R, Stoldt M, Wöhnert J, Görlach M, van Heel M, Brimacombe R (2000). "The 3D arrangement of the 23 S and 5 S rRNA in the Escherichia coli 50 S ribosomal subunit based on a cryo-electron microscopic reconstruction at 7.5 Å resolution". J Mol Biol. 298 (1): 35–59. doi:10.1006/jmbi.2000.3635. PMID 10756104.
- Pei A, Nossa CW, Chokshi P, Blaser MJ, Yang L, Rosmarin DM, Pei Z, Stajich JE (5 May 2009). "Diversity of 23S rRNA Genes within Individual Prokaryotic Genomes". PLoS ONE. 4 (5): e5437. doi:10.1371/journal.pone.0005437. PMC 2672173. PMID 19415112.
- Doris, Stephen M.; Smith, Deborah R.; Beamesderfer, Julia N.; Raphael, Benjamin J.; Nathanson, Judith A.; Gerbi, Susan A. (October 2015). "Universal and domain-specific sequences in 23S–28S ribosomal RNA identified by computational phylogenetics". RNA. 21 (10): 1719–1730. doi:10.1261/rna.051144.115.
- Long KS, Munck C, Andersen TM, Schaub MA, Hobbie SN, Bottger EC, Vester B (9 August 2010). "Mutations in 23S rRNA at the Peptidyl Transferase Center and Their Relationship to Linezolid Binding and Cross-Resistance". Antimicrobial Agents and Chemotherapy. 54 (11): 4705–4713. doi:10.1128/AAC.00644-10. PMC 2976117. PMID 20696869.