3
I have some DICOM medical imaging files downloaded from Cancer Imaging Archive. I can convert them from DICOM to several other formats, but as you'll see the conversion isn't working as expected in most cases.
These are the various conversions I've figured out so far:
- DICOM to netpbm format:
dctopgm8 000005.dcm 000005.pbm - DICOM to pnm format:
dctopnm -byteorder little 000005.dcm 000005.pnm - DICOM to png format:
dcm2pnm +on 000005.dcm 000005.png - DICOM to png format (via ImageMagick):
convert 000005.dcm 000005.png
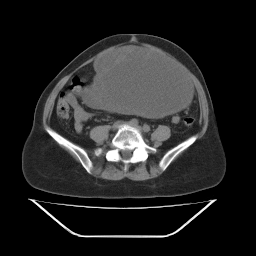
Of those, the .pbm is the only one that seems to give great results. It looks like this:
The .pnm looks like this, which is not quite an inverse image, but somehow looks wrong:
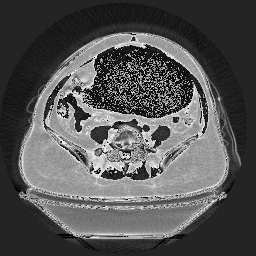
And both of the .png conversions look like this, which is a very washed out image, perhaps due to problems with an alpha channel, gamma, or...?
The problem is I need these to be in PNG, not PBM. And while I could add an additional conversion from PBM to PNG, I'd rather call convert only once and do the full conversion in a single command.
Anyone know what parameters I might be missing in the calls to dcm2pnm or ImageMagick's convert to get the images looking as expected?
Edit: including a sample .dcm image: 000005.dcm

1Why not do it in two steps:
dctopgm8 000005.dcm 000005.pbmandconvert 000005.pbm 000005.png? – Ljm Dullaart – 2019-10-30T09:43:25.310@LjmDullaart Because I'd rather understand what parameters I'm missing so I can do the conversion in a single step. – Stéphane – 2019-10-30T09:45:15.937
I don't really get what the problem is. Your last command is a single conversion command. The other commands, if still necessary, highlight what needs to be done. For a single command
dcmj2pnmseems to provide what you're asking for? – Seth – 2019-10-30T09:53:04.513@Seth I would like to convert the DICOM to PNG format with a single conversion command. – Stéphane – 2019-10-30T09:55:00.647
Already answered here>>>>>>>https://stackoverflow.com/questions/46968678/whats-the-easiest-way-to-convert-a-dicom-image-to-png
– Moab – 2019-10-30T13:36:24.090@Moab No, as you can see above, that technique does not work. The images are not converted correctly. The only one that seems to work correctly is the .pbm, but what I need is .png or .jpg. – Stéphane – 2019-10-30T14:23:17.137
It may not be able to be done in Linux, I see several windows utilities that claim to do it, but this one has to be complied>>>>>>https://support.dcmtk.org/docs/dcmj2pnm.html
– Moab – 2019-10-30T15:00:43.193@Moab, no, that one is part of the usual dcmtk package on Ubuntu, but it doesn't convert the image correctly. See the images I posted in my question. So far, the only one that seems to work correctly is the .pbm converter. – Stéphane – 2019-10-30T15:10:42.727
@Seth I don't know how you can say that. I even included the images that resulted from running those commands. – Stéphane – 2019-10-31T08:37:55.090
Because you haven't. There is a j in that command that isn't in the other. While it's is possible that it will have the same result at least give it a shot. You could also experiment with the input commands for either command and the LUT options. Using verbose and image info might shed some more light on your input and what's going wrong during conversion. – Seth – 2019-10-31T09:25:04.377