Gene expression
Gene expression refers to all the processes involved in converting genetic information from a DNA sequence to make that protein. In prokaryotes, there are just two processes required: transcription and translation. In eukaryotes, there is an additional step, RNA processing (splicing), which intervenes.
This article is only a brief description of the subject and is not intended to give a full explanation.
Check out the "see also" or "references" sections, or Wikipedia's article for more detail.
| Live, reproduce, die Biology |
| Life as we know it |
|
| Divide and multiply |
| Greatest Great Apes |
|
v - t - e |
Transcription
The synthesis of a single-stranded RNA molecule using DNA as a template is referred to as transcription. The enzyme that catalyzes this reaction is known as RNA polymerase. Although the subunit structure and details of the process differ significantly in prokaryotes and eukaryotes, the chemical reaction is identical.
Prokaryotic transcription

Initiation
Bacterial RNA polymerases are composed of multiple subunits. The core enzyme consists of 2 α, β and β′ subunits. This enzyme contains the catalytic activity for RNA synthesis. Association of the core enzyme with the σ subunit is required for initiation of transcription. This complex is referred to as the holoenzyme. The holoenzyme binds to DNA nonspecifically and scans for a promoter sequence. In E. coli, the most common type of promoter contains two conserved sequences centered at -10 and – 35. The σ subunit interacts specifically with these sequences in the double stranded DNA, positioning the active site at +1 (transcription start site). This complex is referred to as the closed complex.
The second step in transcription initiation involves the unwinding of the DNA between the -10 region and the start site of transcription. This is referred to as the open complex.

Elongation
The first two rNTPs complementary to the DNA bind to the active site of RNA polymerase and the first phosphodiester bond is formed. Synthesis is always in the 5’ to 3’ direction and involves the nucleophic attack by the 3’OH on the 5’ phosphate with the release of pyrophosphate. After approximately 8-10 ribonucleotides are added, σ factor is released and RNA polymerase core enzyme moves away from the promoter, synthesizing RNA as it goes.
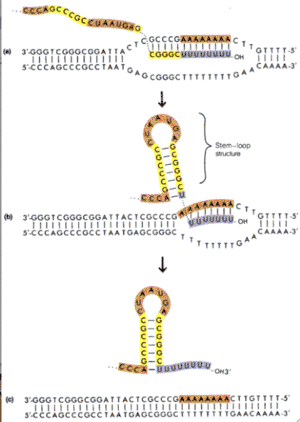
Termination
Transcription continues until a termination signal is reached. The current model is that the RNA dissociates from the DNA – RNA hybrid to form a stem-loop structure. The remaining AU base pairs are not strong enough to keep RNA hybridized with the DNA. Dissociation of the RNA destabilizes RNA polymerases and transcription terminates.
Eukaryotic transcription

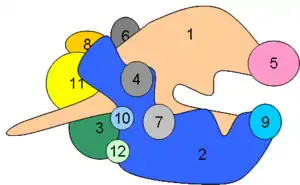
The basic features of RNA synthesis are shared between prokaryotes and eukaryotes; however, transcription in eukaryotes differs in that it is significantly more complex. First, rather than having a single RNA polymerase, eukaryotes have three different RNA polymerases, each of which transcribes a different set of genes. RNA polymerase I transcribes three types of rRNA (the 18S, 5.8S, and 28S species), RNA polymerase II transcribes mRNA, and RNA polymerase III transcribes tRNA and the smallest rRNA (the 5S species). The eukaryotic RNA polymerases consist of between eight and fourteen subunits, with two of them corresponding to the β and β′ subunits of prokaryotic RNA polymerases.
All three eukaryotic polymerases have some homology to the E. coli core polymerase (α2ββ′), but also contain additional subunits not found in prokaryotes. In total, approximately 12-17 subunits (varies with the species) are required for activity in vivo, making eukaryotic polymerases much more complex than prokaryotic polymerases.
RNA Polymerase II
Near the carboxy end of Pol II’s largest subunit a unique region is found (CTD, carboxy terminal domain). This region contains a stretch of 7 amino acids that is repeated between 26 and 52 times (differences in the number of repeats occur in a species specific manner). During transcription initiation several amino acids in the repeat becomes phosphorylated.

Promoters
A typical eukaryotic promoter is located from approximately -40 to +50 relative to the start site (+1). This is referred to as a “core promoter”, with additional regulatory sites being located nearby or at a large distance (enhancers and silencers). Within the core promoter several conserved sequences have been identified. The most prominent (though not found in all promoters) is the TATA motif (5'-TATAAA-3') or TATA box (Goldberg-Hogness box [1]). Located at approximately -30, this highly conserved sequence is easy to identify. However, some promoters are missing the TATA box and through the analysis of these promoters other less conserved sequences have been found. Once is located around +1 and is called the Initiator element (Inr).
Although Pol II is very large and complex, it is unable to neither recognize specific promoter sequences on its own nor catalyze all the steps required for transcription initiation. It requires the help of several factors known as general transcription factors (GTFs). The factors required at most promoters are TFIIA, TFIID, TFIIE, TFIIF and TFIIH.

Holoenzyme
TFIIE, TFIIF and TFIIH associate with RNA Polymerase II and another protein complex (the mediator) to form a holoenzyme. TFIIA and TFIIB are sometimes found together with the holoenzyme, and in other experiments they seem to bind independently. In any event, the holoenzyme is unable to recognize promoter squences and depends on TFIID binding at the TATA box. TFIIA and TFIIB bind to the DNA adjacent to the TATA box and also interact with TFIID.
TFIID
TFIID is composed of a single polypeptide known as the TATA binding protein or TBP, plus approximately 14 additional polypeptides known as TBP Associated Factors or TAFIIs. TBP specifically binds to the TATA box, causing a sharp bend in the DNA. It is this binding of TBP with its associated factors that begins the assembly of the remaining GTFs and the holoenzyme at the promoter.
TFIIE and TFIIH
After binding of the general transcirption machinery to the promoter, TFIIE and TFIIH assist Pol II in unwinding the DNA at the start site of transcription. One of the subunits of TFIIH is a helicase responsible for the unwinding and TFIIE binds to and stabilizes the single stranded DNA.
Another subunit of TFIIH contains a kinase activity responsible for phosphorylating the CTD region of Pol II. Although this activity can be influenced by many factors, in simple systems it is believed to correlate with the initiation of transcription and that movement of RNA polymerase away from the promoter.
Summary of transcription
Although eukaryotic transcription is much more complex than prokaryotic transcription, initiation involves the same basic steps in both:
- Promoter Recognition
- Unwinding DNA around +1
- Synthesis of first few polynucleotides
- Release of polymerase from promoter
In prokaryotes, termination occures at discrete sites and translation can begin even before transcription has terminated. In eukaryotes, transcription continues past the site where poly-A addition occurs and then terminates randomly (frequently 500 bases or so down stream). RNA processing and splicing is completed in the nucleus before transported to the cytoplasm for translation.
Translation
mRNA
mRNA is not a passive player in translation. It is a nucleic acid, and as such contains digital information that can target the mRNA to different parts of the cell, and has a secondary and tertiary structure that can attract the attentions of regulatory proteins.
In eukaryotes, even before mRNA leaves the nucleus, it is extensively modified from the pre-mRNA that was synthesised by RNA polymerase. mRNA is capped at its 5′ end and complexed to cap binding proteins. It is spliced by U-particles, which tag mRNA and the spliced out lariats with different hnRNPs: SR-rich proteins bind exons and hnRNPs bound to excised lariats package them up and mark them for destruction. The 3′ tail of mRNA has a poly-A tail added, which is bound by PABP. All this must occur before export factors binds, and all these items are required for appropriate export from the nucleus.

mRNA can be targeted to specific places in the cytoplasm: the targeting sequences are found in the 3′ UTR (untranslated region).

Targeting is essential to protein function: nerve cells in the hypothalamus (which secrete hormones from their dendrites) target their mRNAs. Vasopressin mRNA is directed to dendrites by a 400 nucleotide long sequence in the 3′ UTR. It appears that PABP, bound to this region, binds to cytoskeletal motors. Other mRNAs are also targeted: calmodulin dependent protein kinase II has a shorter 'Y' element (below), bound by TB-RBP (testis-brain RNA binding protein).
5′---AUG---UAG---AGAAGCCCTATGCT---AAAA---3′
The cytoplasm contains RNAses, and RNA is in any case inherently unstable to hydrolysis. However, different mRNAs have different stabilities: β-globin mRNA in young erythrocytes is very stable; whereas growth factor mRNA is very short-lived (t½ = 30 min). The length of the poly-A tail regulates half-life: it is slowly trimmed off like a time-fuse by an enzyme known as DAN. Some selected mRNAs have poly-A re-added (especially in oöcytes).

Histone mRNA is unusual in that it lacks a poly-A tail and has a much shorter half-life (about 1 hr) than most poly-A mRNAs. Rather than having a poly-A tail, histone mRNA has protective stem-loop structure at its 3′ end. Histone mRNA is rapidly degraded at the end of S-phase (its half life drops to just 12 min).
Some mRNAs are also regulated by endonucleases that chop off the tail wholesale. This is seen in the regulation of the mRNAs of iron metabolism:

- Ferritin sequesters iron.
- Transferrin transports iron into the cell.
- Cytosolic aconitase binds:
- Ferritin mRNA - this blocks ribosomes from binding by covering the RBS;
- Transferrin mRNA - this blocks the site of action of these endonucleases.
Translation
Transcription is 'easy': DNA and RNA are different 'dialects' of the same language. Translation is more 'difficult': it requires a code book (the genetic code) that converts the nucleic language to that of proteins.
Transcription reads the template DNA strand 3′→5′. RNA polymerase synthesises mRNA 5′→3′. Translation reads the mRNA 5′→3′. The ribosome synthesises protein NH3+ → COO−. It should be obvious, but note that the promoter is not the same thing as the start codon; nor is the RNA polymerase terminator the stop codon. There are generally untranslated regions (UTRs) at both ends of an RNA transcript, even in bacteria.
Translation requires the cooperation of:
mRNA, tRNA (and aminoacyl tRNA synthetases), rRNA, Initiation, elongation and termination factors. Translation has a low error rate: 10−4 for amino acids, and a speed of about 3 amino acids −1 (eukaryotes) to 20 amino acids s−1 (prokaryotes). These parameters are very similar (if measured per nucleotide) to transcription. Translation occurs on the ribosome.
Ribosomes consist of two subunits: a larger one (the LSU) and a smaller subunit (the SSU). Both subunits contain RNA (the larger contains two or three, the smaller just one) and large numbers of proteins (which are mostly structural). The large subunit contains the active site for peptide bond formation, which is a ribozyme (catalytically active RNA) called peptidyl transferase.

The small subunit initially binds to mRNA like a latch, and also binds tRNAs at the A(minoacyl), P(eptidyl) and E(xit) sites. It controls information flow, whilst the large subunit controls the chemistry. Both help form the groove in which mRNA binds. The E, P and A sites are formed by both ribosomal subunits: the tRNAs interact with mRNA in the SSU P and A sites, whilst the synthesis of protein occurs in the P and A sites of the LSU.
Like transcription, translation may be divided into the processes of initiation, elongation and termination. Each step requires the action of several protein factors, RNAs and GTP. It also requires the 'forgotten' part of translation, the aminoacyl tRNA synthetases:
Aminoacyl tRNA synthetases answer the question: "How is tRNA associated with the correct amino acid?" The synthetase selects tRNA by its codon and/or other bases, and attaches the correct amino acid. There are usually just 20 synthetases, many of which must be able to recognise more than one tRNA, so they cannot discriminate purely on the basis of the anticodon.
Amino acid + ATP → Aminoacyl-AMP + PPi.
Aminoacyl-AMP + tRNA → Aminoacyl-tRNA + AMP.
Initiation
Initiation in eukaryotes starts with the formation of the eukaryotic 43S pre-initiation complex:

eIF1A blocks the A site. This is necessary to prevent tRNAs from binding inappropriately. This blockage forces eIF2-GTP to recruit the special initiator tRNAimet to the P site. Only tRNAimet can bind a naked SSU. eIF3 prevents binding of the SSU to the LSU. The combination of the SSU with the three initiation factors and tRNAimet is termed the 43S preinitiation complex. The fourth initiation factor, eIF4A, which is a helicase, unwinds any hairpin loops in the mRNA, exposing the start codon (AUG).

The 43S complex binds to the mRNA and finds the first AUG and eIF2 then hydrolyses GTP, releasing the initiation factors and allowing the LSU to bind and form the eukaryotic 80S initiation complex.
Some mRNAs require additional factors to ensure correct expression at the ribosome: in picornaviruses (such as polio and hepatitis) the VPG protein binds the 5′ end (which is not capped) and directs the ribosome to the correct AUG start codon.
In bacteria, IF1, IF2 and IF3 play similar roles to eIF1, eIF2 and eIF3, and tRNAifmet is always the first tRNA. In both eukaryotes and prokaryotes, the special methionine is usually cleaved off the final polypeptide product. Furthermore, in bacteria, polycistronic mRNAs (which have several start codons) have Shine-Dalgarno sequences c. 5 nt to 5′ of the AUG start codons. This binds the anti-Shine Dalgarno sequence at the end of the 16S SSU rRNA, ensures that the ribosome binds at the correct place(s), not at an out-of-phase codon or an internal methionine.
mRNA: 5′-----AGGAGG-----AUG----3′
rRNA: 3′-…-auUCCUCCacuag---5′
Elongation
The A and P sites are so close on the ribosome that tRNAs must fit contiguous codons. tRNAs are brought in by EF-Tu (bacteria) or EF-1 (eukaryotes), which hydrolyses GTP in so doing.

The second elongation factor, EF-G (EF-2), shunts the ribosome along, again using GTP.

This movement brings amino acyl and peptidyl groups together in the active site, catalysing peptide bond formation, and the transfer of the peptidyl group from one tRNA to the next.

The de-charged tRNA diffuses out of the ribosome via the E site. Note that the nascent polypeptide chain is always covalently attached to the acceptor stem of the tRNA in the P site.

Peptide bond formation is performed by peptidyl transferase, whose active site contains an adenine ring from the 28S rRNA in large subunit. This performs and acid/base catalysis, just like histidine does in many enzymes. The antibiotic puromycin inhibits ribosomes by mimicking tRNAtyr and preventing the normal transfer of the nascent peptide chain from the tRNA in the P site transfers onto the tRNA in the A site.
Termination
Stop codons are bound by cytoplasmic release factors, which add water to tRNApeptidyl, cleaving off the polypeptide.

There are two release factors in prokaryotes (RF1 and RF2), but just one in eukaryotes (eRF1), which I think must be the only case where eukaryotes have evolved a simple, elegant system ☺ RFs mimic tRNA in shape, but are made of protein, not RNA.
Prokaryotes also have a special release factor called tmRNA, which provides a template for stalled ribosomes (ones that have stopped translating without meeting a stop codon, maybe because mRNA has been released too early by RNA polymerase). The tmRNA contains a template for an 11 amino acid tag, which marks these abortive proteins for destruction.
Two of the eIF4 proteins (of which there are several besides the IF4A helicase), eIF4E and eIF4G bind the mRNA cap to the PABP on the tail. This means that ribosomes form coiled structures on bound mRNA. Something similar also occurs in prokaryotes. These 'polyribosomes' increase the efficiency of translation, since less mRNA is needed (the ribosomes are only 80 nt apart, so many can simultaneously translate a single mRNA), and also allow efficient re-recruitment of terminated ribosomes back onto the RBS.

Many antibiotics work on the ribosome or other aspects of transcription or translation:
| Prokaryote | Eukaryote | |
|---|---|---|
| mRNA synthesis | Actinomycin-D,Rifampicin | Actinomycin-D,α-Amanitin |
| Binding tRNA | Tetracycline | |
| Initiation complex | Streptomycin | |
| Peptidyl transferase | Chloramphenicol | Anisomycin |
| Translocation | Erythromycin | Cycloheximide |
| Premature release | Puromycin | Puromycin |
Eukaryotic RNA processing
Splicing
Transcripts in eukaryotes are c. 10 000 nt, but polypeptides are 400 aa = 1200 nt. 80% of transcript is not translated, i.e. pre-mRNA is generally 7800 bases longer than a typical 1200 bp mRNA. What happens to all this excess RNA? Eukaryotic genes (and some Archaeal gene) contain internal 'junk' (discovered in 1977). Exons (which are expressed, and exit the nucleus) are separated by introns (100 - 100 000 nt long), which are removed from the pre-mRNA and destroyed without ever leaving the nucleus. The size of introns is species-specific: yeasts have very few, some human genes can have 50 in a single gene! Typically there are four introns within five exons, but the introns (as mentioned above) are generally much larger than the exons. Introns must be spliced out from between the exons before the mRNA can leave the nucleus.
5′---E1---I1---E2---I2---E3---I3---E4---I4---E5---3′
As mRNA is transcribed, it is complexed by five small nuclear ribonucleoproteins (snRNPs, also known as 'U' particles).RNAse treatment of pre-mRNA degrades pre-mRNA into heterogeneous ribonucleoprotein fragments. These proteins have a common RNA binding motif (below). Some of these hnRNPs are U-proteins, directly involved in splicing out introns, others are important for labelling introns and exons.
H3N+---{KR}G{FY}{AG}{FY}VX{FY}---…---COO−
Sufferers of systemic lupus erythrematosus (SLE) produce autoimmune anti-snRNP antibodies, which can be used to reveal islands of snRNP in nucleus, which are known as Cajal bodies.
Splice sequences are quite variable across species, but in general:
Exon1 ends AG (the donor site). There must be internal A in branch site, which reacts chemically with the donor junction to effect the splice. Exon2 starts G (the acceptor site). 5′ donor branch acceptor 3′
5′---AGGURAGU---CURAYY---YNCAGG---3′
5′---AGG---3′
The intron sequence is spliced out of the transcript to generate the processed mRNA. This is the human consensus sequence.
The U-particles combine with one another in a variety of ways to for spliceosomes. In the most typical splicing reaction, U1 binds the 5′ donor. U2 then binds the 3′ acceptor and bridge sequence. U4 and U6 act together to form a lariat, and then U5 ligates the exons together. Note that U2, U5 and U6 remain attached to the lariat in U-catalysed splicing. This helps mark the lariat for degradation.
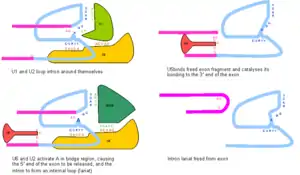
Tetrahymena (a ciliate) and some Archaea have self-splicing introns. The snRNP system described above probably evolved from (type-II) self-splicing systems. Note that there are several other varieties of splicing, using the U particles whose existence you might be wondering about.

The donor and acceptor splice junctions are correctly paired off in multi-intron genes because splicing is concurrent with transcription. Some specificity may also be allowed by the hnRNPs and SR-rich proteins that bind to the pre-mRNA. Capping, splicing and poly-A factors are associated with the tail of RNA polymerase II: the enzyme complex is a factory, both producing mRNA and processing it ready for export.
Splicing seems to be an expensive waste of time for the most part; however, under certain circumstances, it has advantages in producing more than one gene product from a single stretch of DNA. In B-lymphocytes, early in development, the cell produces membrane-tethered immunoglobulin (Ig) receptors by transcription and splicing of the entire gene:
Long transcript:
5′---Ig---donor---stop---acceptor---hydrophobic---stop---3′
After splicing:
5′---Ig----hydrophobic----stop----3′
Protein:
H3N+-Ig-hydrophobic-COO−
The longer transcript splices out the first stop codon, producing an antibody with a hydrophobic carboxy terminus, which allows it to be tethered into the cell membrane. However, after the cell has recognised an antigen, it begins to produce soluble antibodies instead. Recognition of antigen increases the amount of CStF in the cell, producing a shorter transcript of the gene, which lacks the intron acceptor sequence:
Short transcript:
5′---Ig---donor---stop---3′
No splicing, because of lack of acceptor junction, so the protein is:
H3N+-Ig-COO−
The shorter transcript produces a protein with no hydrophobic peptide, which is plasma-soluble.
We've mostly been speaking about mRNA modification, but the other types of RNA are even more heavily modified (edited) than mRNA…
tRNAs are c. 80 bases long, and in eukaryotes, there are at least 31 in the cytoplasm, 30 in chloroplasts and 22 in mitochondria. All tRNAs have an acceptor stem with a 3′ CCA sequence, which (covalently) binds amino acids. They all also have a triplet anticodon, which (transiently) binds mRNA. The general structure is often termed a 'cloverleaf', which is true of the secondary structure, but somewhat misleading for the tertiary.

Modification of the tRNA anticodon produces gibberish proteins. Chemical modification of tRNAcys to tRNAala also produces non-functional proteins. The anticodon/amino-acid pairing is essential to correct translation of mRNA.
tRNA contains unusual nucleosides. Inosine (needed for wobble). Pseudouridine (only in Eukaryotes and Archaea). Dihydrouridine. Thymine (unusual for RNA). Wybutosine (found just after the anticodon).
Chemical modification of tRNAs is performed by proteins, unlike mRNA splicing. One major effect of modification is to allow wobble. Wobble is the non-Watson-Crick pairing at the third base of a codon, which means that fewer anticodons (tRNAs) are needed than the 64 possible codons: 64 codons. 20 amino acids. 30 is the compromise made.
There is generally more wobble in bacteria (see bracketed bases below). Mitochondria even have wobble at the second base of the codon.
| Wobble codon base | Possible anticodon bases |
|---|---|
| U | A), G, I |
| C | G, I |
| A | U, (I) |
| G | C, (U) |
In trypanosome mitochondria, guide RNA (gRNA) guides excision and addition (insertion/deletion or indel) of bases (mostly U) to mRNA.
gRNA above; mRNA below:
3′---UU AUCUCAACCGACCA---5′
5′---AAGUAGAG~~GGCUGGU---3′
Note that the is G displaced and the AA causes stretching, so the edited RNA looks like:
5′---AA|UAGAGUUGGCUGGU---3′
With the G excised and UU inserted in the stretch.
Bibliography
- Stryer, Lubert. Biochemistry, 4th ed. New York: W. H. Freeman and Company, 1995
- Tijian, Robert. "Molecular Machines that Control Genes." Scientific American 272 (1995): 54–61.
References
- Lifton RP, Goldberg ML, Karp RW, Hogness DS. (1978). The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 42, 1047-1051. PMID 98262